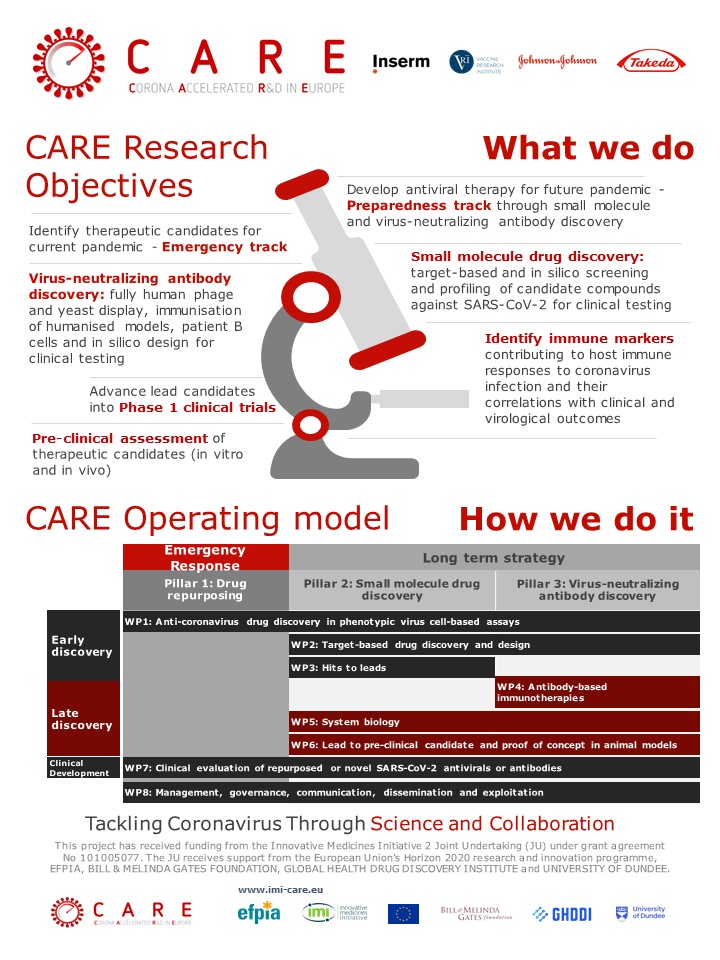
Why did Utrecht University choose to get involved in CARE?
The Virology Section has 50+ years of experience in coronavirus biology and possesses extensive expertise in both human and animal coronaviruses. They have particular expertise in virus structure, virus-receptor interactions, virus entry mechanisms, genome replication and virus-host interactions. The CARE project enabled them to quickly contribute their knowledge with many other research groups, while also benefitting from enhanced collaboration with the CARE partners.
What has Utrecht University delivered for CARE?
UU’s contributions span across various innovations and findings in Work Packages (WPs) 1, 2, 4 and 5. Overall, their efforts aim to identify potential antiviral drugs, peptides, nanobodies and antibodies, to understand their mechanisms of action, and to explore host factors involved in coronavirus infection, which could inform the development of effective therapies against SARS-CoV-2 and other related coronaviruses.
In WP1, they evaluated a large number of small molecules for antiviral activity against SARS-CoV-2 and other coronaviruses, characterized antiviral effects and performed Mechanism of Action studies to reveal their molecular mechanisms.
In WP2, they developed macrocyclic peptide inhibitors and nanobodies targeting the Spike protein of SARS-CoV-2. The team studied the site in Spike that is targeted by the macrocyclic peptide using cryo-Electron Microscopy (cryo-EM). This revealed that the peptide binds to a conserved region of the SARS-CoV-2 spike protein that has hitherto not been exploited by antibodies or small molecules and has not been mutated in any of the variants of concern.
In WP4, they established assays, reagents and structural biology workflows for the assessment and characterization of human neutralizing antibodies (nAbs) against SARS-CoV-2 and variants. The team generated bispecific antibodies to enhance potency and breadth of nAbs against SARS-CoV-2 and related coronaviruses. They also developed broad-spectrum nAbs against porcine deltacoronavirus (PDCoV), an emerging coronavirus with pandemic potential. Finally, they isolated and characterized human antibodies that target host receptor aminopeptidase N (APN). These antibodies are highly resistant to viral escape and can be employed for coronaviruses that share the same receptor for entry, increasing our pandemic preparedness against newly emerging coronaviruses.
In WP5, they determined changes in the metabolome in coronavirus-infected cells and identifying essential host factors for coronavirus infection via genetic CRISPR-cas9 screens. In follow-up experiments, they are investigating the physiological importance of the virus-induced changes in metabolism and the role of the identified host factors for efficient virus replication and/or evasion of infection-limiting host antiviral responses.
For more information about the different work packages, please click here
Who is working in the CARE team at Utrecht University?
The Utrecht University team is led by two Principal Investigators: Professor Dr Frank van Kuppeveld who studies the interaction between viruses and their host; and Dr Berend Jan Bosch (Associate Professor) who studies virus-receptor interactions and cell entry mechanism of membrane-enveloped viruses, particularly coronaviruses.
During the CARE project, Dr Daniel Hurdiss, a structural virologist, was promoted from a post-doctoral researcher to Assistant Professor. He is a structural virologist who, among others, uses cryo-EM to study the 3D structure of viral proteins and understand their functional implications.